Physical Molecular Biology Section
Computer-generated
Image of a Model Chromatin Fiber
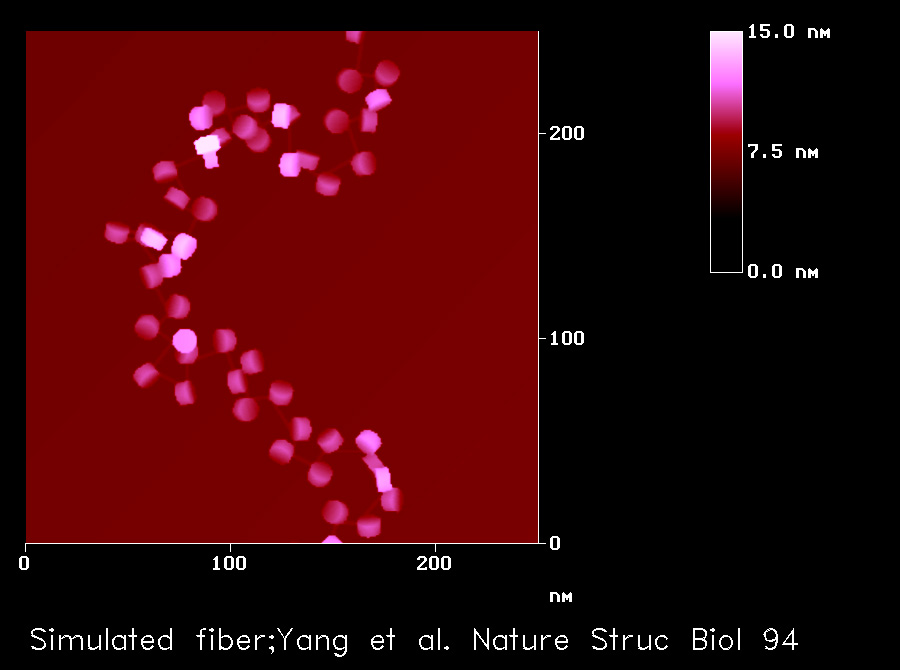
The nucleosome core is modelled as a disc of 5.5 nm high and 11 nm in diameter, which includes 1.75 turns of DNA wrapped around a histone octamer in a left-handed fashion with a
pitch of 28.6 Å. The exit angle of the DNA is determined by the tangent at the point it leaves the nucleosome. The linker DNA is assumed to be straight between nucleosomes. The length of the linker DNA varies between 51 and 73 bp, which is determined using a uniform deviate random-number algorithm. The volume exclusion effects aretaken into account by assuming a spherical hard-core potential with a radius of 10.2 nm around each nucleosome. A smaller potential sphere leads to non-fiber-like structures. A helical repeat length of 10.15 bp per turn is used for the DNA wrapped around the histone octamer, whereas 10.40 bp per turn is used for the linker. To generate the image,each nucleosome in the model fibre is projected onto a plane without changing its orientation. The plane is chosen such that the sum of its distances to all the nucleosomes in the fibre is a minimum.
Yang, G., Leuba, S. H., Bustamante, C., Zlatanova, J. and van Holde, K.
Role of linker histones in extended chromatin fibre structure. Nat Struct Biol. 1(11) 761-763 1994
